Project Summary
Distribution of Acyl-homoserine-lactone Autoinducer (AHL) in the Gastrointestinal Tract of Ruminants and Correlation to Season, E. coli O157:H7 Prevalence, and Diet
- Principle Investigator(s):
- Tom S. Edrington and Vanessa Sperandio
- Institution(s):
- U.S. Department of Agriculture, Agricultural Research Service
- Completion Date:
- May 2008
Background
Enterohemorrhagic E. coli (EHEC) O157:H7 is responsible for major outbreaks of bloody diarrhea and hemolytic uremic syndrome (HUS) throughout the world. EHEC causes an estimated 73,000 illnesses, 2,000 hospitalizations, and 69 deaths in the United States alone each year. Cattle are natural reservoirs for this bacterium and typically appear non-symptomatic while shedding this pathogen into the environment. A previous study performed a signature tagged mutagenesis study (STM) to identify EHEC genes necessary for colonization of cattle. In this study the researchers identified 59 EHEC genes required for the intestinal colonization of cattle. Among these genes were the genes from the locus of enterocyte effacement (LEE), necessary for EHEC adhesion to human and cattle intestines, and sdiA, encoding the LuxR-homolog SdiA in E. coli. LuxR is the sensor for the quorum sensing (QS) system in Vibrio fisheri and the LuxR/I system was the first one to be described in Vibrio fischeri. Quorum sensing is a bacterial cell-to-cell signaling mechanism through which bacterial cells assess the density of their population. These bacteria secrete hormone-like compounds, usually referred to as autoinducers that, when at threshold concentrations, interact with transcriptional regulators to drive bacterial gene expression. The quorum sensing system in Vibrio fischeri consists of two proteins, LuxI, which is responsible for the production of the acyl-homoserine-lactone (AHL) autoinducer and LuxR, which is activated by this autoinducer to increase transcription of its target genes. Since this initial description, homologues of LuxR-LuxI have been identified in other bacteria and in all of these LuxR-LuxI systems, the bacteria produce an acyl-homoserine lactone autoinducer (AHL), which binds to the LuxR protein and regulates the transcription of several genes involved in a variety of phenotypes. Escherichia coli and Salmonella have a LuxR homologue, SdiA, but do not have a LuxI gene, and do not produce AHL. The precise role of SdiA in QS was elusive for several years until it was reported that SdiA is not sensing an autoinducer produced by E. coli itself, but rather AHL produced by other bacterial species. However, no targets for SdiA regulation are known in EHEC. The fact that a functional SdiA is necessary for EHEC to successfully colonize the intestinal tract of cattle is especially intriguing given previous observations that the rumen contains bacteria that produce the AHL signals. A sdiA mutant was generated and observed that the AHL signals produced by bacteria within the rumen of cattle, repress expression of the LEE genes (also necessary for colonization). Gene array experiments were also performed, and other putative novel adhesions in EHEC were found that are also under the control of SdiA and AHL signaling. Taken together, preliminary data, combined with the previous literature, suggest that AHL in the rumen of cattle are involved in repressing expression of genes in EHEC that are necessary for the colonization of cattle. Consequently, in this proposal the researchers wanted to expand our knowledge of this system by testing whether seasonality or diet may play a role in the presence of these signals in the ruminant. In addition, it will be assessed whether there is any correlation between increase or decrease in AHLs in the rumen of cattle and fecal shedding of EHEC. Finally, in light of the recent research suggesting the rectal-anal junction is the primary site of EHEC colonization in cattle, it will be investigated whether there are any AHL present in other segments of the bovine gastro-intestinal tract, or if their presence is restricted to the rumen.
The stated objectives for this work were:
- Determine prevalence and concentrations of AHL in luminal contents collected from the rumen, small intestine, colon and rectum of feedlot cattle at slaughter.
- Determine if season influences AHL concentrations by sampling feedlot cattle in the spring, summer, fall and winter.
- Examine populations of E. coli O157:H7 (qualitatively and quantitatively) in above luminal content samples and determine if correlated to AHL concentrations.
- Utilizing growing lambs as an inexpensive cattle model, evaluate concentrations of AHL in luminal contents throughout the gastro-intestinal tract of lambs fed forage- or grain-based diets.
Methodology
Sample collection. All samples were collected from feedlot cattle at a single slaughterhouse in the southwestern United States. Four collections, encompassing the four seasons, were made in May, August and October of 2007, and January 2008. Luminal content samples were aseptically collected from the rumen (1000 ml) and rectum (50 ml) from 60 head at each seasonal collection. During the spring collection, samples were also collected from the cecum and small intestine and analyzed, however these samples were not included in future collections due to the absence of AHL in these GIT segments. Samples were shipped on dry ice to the laboratories in Dallas (AHL analysis) or College Station, TX (E. coli O157:H7 culture) for processing the following day as described below.
Sheep study. Fourteen crossbred weaned lambs (avg. BW = 25 kg) were purchased and transported to the livestock facilities at the USDA laboratory in College Station, TX. Upon arrival, lambs were ear-tagged, vaccinated for enterotoxemia, weighed and randomly assigned to treatment: Forage (90% grass hay, 10% grain/mineral supplement) or Concentrate (80% commercial sheep feed, 10% grass hay). Following a two-week adaptation period to diet, all sheep were individually penned and inoculated with a cocktail of two strains of E. coli O157:H7 made resistant to different antibiotics (rifampicin, novobiocin, naladixic acid). Ten mL of each strain [933 (4.9 x 109/mL) and 6058 (5.3 x 109/mL)] were administered to all lambs via oral gavage. Fecal samples were collected daily for seven days from each lamb for serial quantification of each of the inoculated strains of EHEC. All lambs were humanely euthanized on day 8 post-inoculation and necropsied. Luminal contents and tissue were aseptically collected from the rumen, ileum, cecum, spiral colon and rectum. Luminal contents were serially diluted for quantification of the challenge strains of EHEC and assayed for prevalence and concentrations of AHL. Tissue samples were qualitatively cultured for the challenge strains of EHEC.
Detection of AHL in ruminal fluid and luminal contents. AHL were extracted from ruminal fluid and luminal contents using dichoromethanol, and subsequently tested in a reporter strain for their presence. This reporter strain is a strain of Agrobacterium tumefaciens that has a gene regulated by AHL fused to a gene encoding β-galactosidase, that in the presence of AHL and its substrate (X-Gal), turns blue. The AHL extracted were further defined through thin layer chromatography (TLC) to identify their chemical composition.
Assessment of LEE gene regulation by AHL. A portion of the AHL extracted as described above were incubated with EHEC and used to compare expression of the LEE genes in EHEC with and without AHL. Expression of the LEE was assessed through real-time RT-PCR. RNA from these cultures was extracted using a RiboPure – Bacteria RNA isolation kit following manufacturer’s guidelines. Real-Time RT-PCR was performed in a one-step reaction using an ABI 7500 sequence detection system. For each 20 μl reaction, 10 μl 2X SYBR master mix, 0.1 μl Multi-scribe reverse transcriptase (Applied Biosystems), and 0.1 μl RNase inhibitor were added. Amplification efficiency of each of the primer pairs was verified using standard curves of known RNA concentrations. Melting curve analysis was used to ensure template specificity by heating products to 95°C for 15 s, followed by cooling to 60°C, and heating to 95°C while monitoring fluorescence. Once amplification efficiency and template specificity are determined for each primer pair, relative quantification analysis was used to analyze the unknown samples using the following conditions for cDNA generation and amplification: 1 cycle at 48°C for 30 min., 1 cycle at 95°C for 10 min., 40 cycles at 95°C for 15 s and 60°C for 1 min. The rpoA (RNA polymerase subunit A) gene was used as the endogenous control.
Bacterial culture, isolation and enumeration methods. Luminal content samples from feedlot cattle were collected and shipped immediately to our laboratory in College Station, TX for qualitative and quantitative analysis of E. coli O157:H7 using a spiral plating method developed at MARC, Clay Center, NB (quantitative) and an immunomagnetic separation technique (qualitative). For the sheep study, fecal samples and luminal contents (1 g) were serially diluted in sterile phosphate-buffered saline and plated on each of two agars containing antibiotics used to mark each EHEC strain. Colonies exhibiting typical EHEC morphology were manually counted following incubation. Tissue samples were rinsed with sterile water, enriched (tetrathionate broth, 24 hr, 37°C) and a portion of the enrichment plated on each of the selective agars for qualitative culture of the challenge strains.
Statistical analysis. All data from the sheep study were analyzed using SAS Version 8.02 (SAS Inst. Inc., Cary, NC, USA). Daily fecal shedding data was analyzed using the PROC Mixed procedure with treatment, day and treatment x day interactions included in the model. The incidence of E. coli O157:H7 positive tissues and luminal contents were subjected to Chi-square analysis using the Proc Freq procedure. Differences among means were considered significant at a 5% level of significance.
Findings
Expression of LEE is required for EHEC to colonize the GIT. Ten rumen samples from the spring collection were examined and all were found to repress LEE expression (Table 1). This finding is consistent with the general understanding of EHEC location and colonization within the GIT. Therefore, it is likely that the presence of AHL in the rumen of cattle represses LEE expression and thereby reduces EHEC prevalence in the rumen, while in the lower GIT, pH does not favor AHL and LEE is not repressed, allowing EHEC to colonize and persist.
Implications
Preliminary data and previous literature suggest that AHL produced by non-EHEC bacterial species in the rumen of cattle are involved in repressing expression of genes in EHEC that are necessary for colonization of the GIT. There appears to be a weak correlation to AHL and EHEC prevalence, particularly in the winter data from feedlot cattle. Taken together, these findings indicate that AHL, or more importantly LEE expression, may be as or more important than the development of acid resistance by EHEC for the colonization of the lower GIT.
Table 1. Prevalence of E. coli O157:H7 [following direct plating (DIR) or enrichment (ENR) and AHL and repression of the LEE gene expression in feedlot cattle at slaughter. Luminal content samples were collected seasonally from the rumen, small intestine (SI), cecum and rectum (nd = not determined).
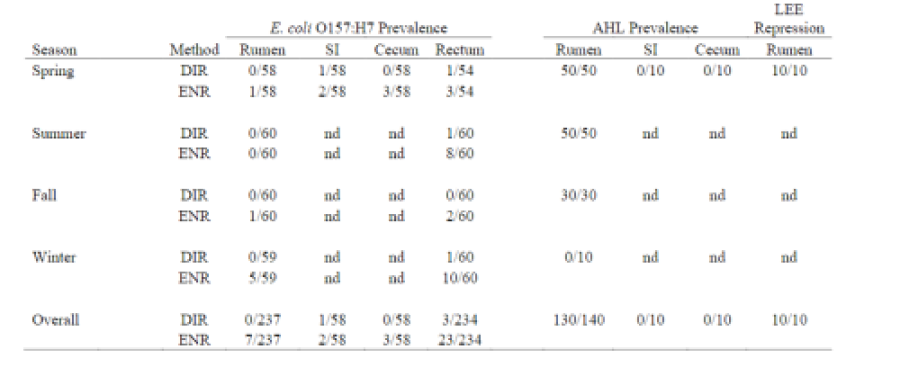
Table 2. Daily fecal shedding [cfu (log10)/g feces] of two strains of E. coli O157:H7 (6058 and 933) in experimentally inoculated sheep fed wither a forage- or grain-based diet.
|
|
Forage |
Grain |
||
|
Day |
6058 |
933 |
6058 |
933 |
|
1 |
5.16 |
5.39 |
5.97 |
5.83 |
|
2 |
3.69 |
3.44 |
3.68 |
3.51 |
|
3 |
2.73 |
2.89 |
2.78 |
3.51 |
|
4 |
1.7 |
2.46 |
1.76 |
2.61 |
|
5 |
1.44 |
3.42 |
1.97 |
1.77 |
|
6 |
1 |
3.02 |
1 |
1.88 |
|
Average |
2.61 |
3.44 |
2.86 |
3.19 |
Table 3. Tissue and luminal content enrichments positive for E. coli O157:H7 (strain 933) in experimentally inoculated sheep.
|
|
Diet |
|
|
|
Item |
Forage |
Grain |
P>F |
|
Tissue enrichments (% positive) rumen |
|
|
|
|
Rumen |
50 |
14.3 |
0.16 |
|
Cecum |
66.7 |
28.6 |
0.17 |
|
Ileum |
83.3 |
57.1 |
0.31 |
|
Colon |
50 |
57.1 |
0.8 |
|
Rectum |
16.7 |
57.1 |
0.13 |
|
Luminal content enrichments (% positive) |
|
|
|
|
Rumen |
100 |
100 |
1.0 |
|
Cecum |
33.3 |
42.9 |
0.72 |
|
Ileum |
33.3 |
28.6 |
0.85 |
|
Colon |
16.7 |
28.6 |
0.61 |
|
Rectum |
16.7 |
28.6 |
0.61 |

Figure 1. AHL prevalence in ruminally cannulated cattle consuming a forage or grain (concentrate) based diet.
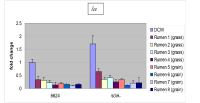
Figure 2. Repression of LEE gene expression in rumen fluid from ruminally cannulated cattle consuming a forage or grain-based diet.
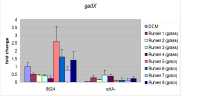
Figure 3. Repression of the gadX gene expression required for the development of acid resistance in rumen fluid from ruminally cannulated cattle consuming a forage or grain-based diet.