Project Summary
Gene Expression Response of Salmonella to Simulated Multiple-hurdle Carcass Processing Interventions
- Principle Investigator(s):
- Dayna M. Brichta-Harhay, Tim P. L. Smith and Gregory P. Harhay
- Institution(s):
- U.S. Department of Agriculture, Agricultural Research Service
- Completion Date:
- May 2010
Background
The foodborne pathogen Salmonella accounts for an estimated 1.3 million cases of
gastroenteritis per year in the United States and is a common contaminant of cattle hides at slaughter. Cattle hide contamination may be transferred to carcasses during the dressing process. In response to this problem, many beef processing plants have implemented sequential carcass interventions aimed at eliminating the presence of pathogens on final carcasses. Numerous studies have demonstrated the efficacy of these multiple-hurdle processing interventions at reducing the presence of pathogens on final carcasses.
Despite the efficacy of these interventions, the low-level prevalence of pathogens on final carcasses and in beef products persists, and there is growing concern that pathogens may be adapting to survive interventions rather than simply avoiding their effects due to attachment in “hard to reach” locations on the carcass surface. If this is true, then survivors of the carcass intervention process may emerge as “resistant organisms” (with “resistant” meaning “resistant to environmental stress”) in the final product. This concern is fueled in part by the observation that many pathogens exhibit an adaptive response when exposed to harsh environmental conditions. Once exposed to an environmental stressor, for example “acid shock” or “heat shock,” they adapt to withstand prolonged exposure to that stressor. The acid tolerance response of Salmonella is a well-studied aspect of this phenomenon. Moreover, these studies have shown that acid adaptation in Salmonella induces cross-protection against other stresses including heat, salt and oxidative stress.
Studies of the physical response of Salmonella to various environmental stressors have typically involved the use of classical microbiological techniques and were limited to Salmonella serotypes frequently associated with human salmonellosis, namely Typhimurium and Enteriditis. Consequently, there is little information on the response of a broader spectrum of important Salmonella serotypes to environmental stressors, such as those imposed by carcass interventions commonly used in beef processing plants, including hot water, organic acid rinse and cold temperature exposure. Here the researchers characterized the gene expression profile of a multi-drug resistant (MDR) Salmonella Newport strain in response to beef carcass processing interventions. The results of this study represent the most comprehensive picture to date of the molecular strategies that Salmonella employs to survive these interventions.
The stated objectives for this work were to investigate changes in gene expression by Salmonella in response to simulated multiple-hurdle carcass processing interventions, with the aim of understanding the adaptive response to these interventions.
Methodology
In order to address the knowledge gaps described above, the researchers examined the gene expression response of Salmonella when exposed to simulated carcass processing interventions. The strain selected for this study was a multi drug-resistant Salmonella enterica serotype commonly associated with cattle, S. Newport. This strain had been previously isolated from a post-intervention cattle carcass. The genomic DNA sequence of this strain was determined using a Roche GS-FLX pyrosequencing instrument. For the gene expression response studies, the S. Newport strain was cultured in nutrient rich conditions, and then exposed to sequential environmental stressors, including hot water (80°C), 2% lactic acid (55°C) and prolonged exposure to cold temperatures (4ºC). The gene products (messenger RNA or mRNA) expressed in response to these stimuli were isolated, converted into DNA and then sequenced, again using the Roche 454 high sequencing platform. Gene expression was measured by mapping cDNA sequences back to an annotated Newport complete genome sequence.
Findings
This study has demonstrated the utility of RNA-sequencing methods for the purpose of characterizing the gene expression response of Salmonella to simulated beef processing interventions. Mapping the cDNA sequence reads back to the reference genome showed that intervention exposure resulted in the increased expression (greater than 2-fold in comparison with untreated cells) of 761 genes (~16% given 4,613 genes examined). Genes demonstrating the greatest fold change included those with noted roles in the heat shock response, DNA repair, two component response regulator systems including those that control outer membrane lipopolysaccharide (LPS) modification and ATP-binding cassette (ABC) transport systems (Figure 1). Also noteworthy, were a number of differentially expressed genes that were found to be involved in multiple metabolic pathways (Table 1). Examination of these data showed that genes involved in cysteine and methionine metabolism, fatty acid biosynthesis/metabolism, histidine utilization, and DNA repair/synthesis were highly upregulated in response to simulated intervention exposure. This observation represents a potentially significant result, as the genes identified here, and the proteins they encode, represent strong targets for novel interventions. Targeting these “lynchpin” metabolic steps would likely result in wide-ranging deleterious effects for the pathogen. By characterizing and comparing the gene expression profiles of Salmonella Newport in response to the environmental stimuli described, this study has highlighted a number of genes, pathways and regulatory elements that may be instrumental in enabling this organism to survive current intervention strategies. Further studies aimed at targeting these genes and pathways could lead to the development of novel antimicrobial agents, designed to prevent Salmonella entry into the beef food chain.
Implications
By examining gene expression profiles of Salmonella serotypes responding to the most commonly used interventions, there have been specific microbial genes and pathways identified that likely enable Salmonella to survive processing interventions. At minimum, the results of this study have increased understanding of the adaptation mechanisms that Salmonella employs in response to commonly used carcass processing interventions. The data generated in this study may provide information for developing modified processing hurdles that will aid the beef industry in more efficiently removing foodborne pathogens from beef processing environments.
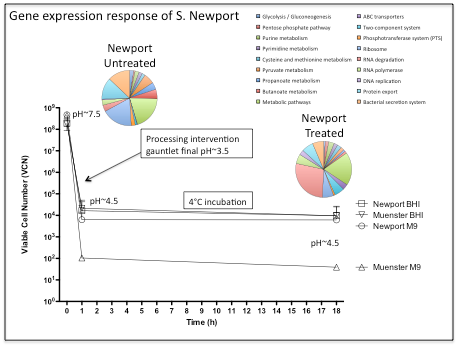
Figure 1. Gene expression responses of S. Newport to simulated carcass interventions. The graph depicts the viable cell number (VCN) of Salmonella in response to exposure to simulated multiple hurdle interventions, including: 80°C (30 sec), 35°C (pH 3.5) (10 min), 55°C 2% lactic acid (final pH ~3.5) (30 sec), 35°C (pH 3.5) (10 min), and chill (4°C) 18h (~4.5). The pie charts indicate the changes in S. Newport gene expression (color code listed) when exposed to the environmental stressors described. Treated Newport: gene expression in cultures exposed to multiple hurdle interventions; Untreated Newport: gene expression in the control Newport reference cultures.
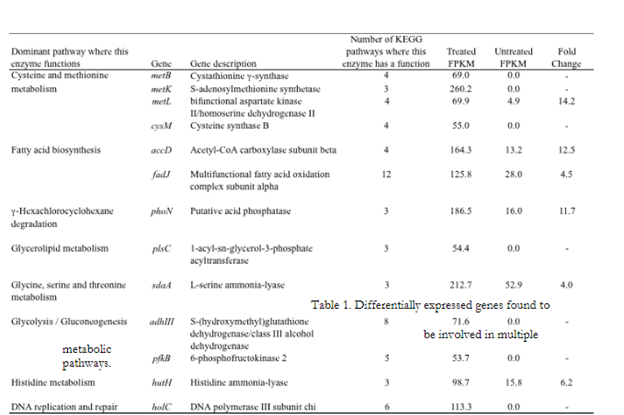
Figure 2. These genes and their protein products represent potentially novel targets for beef processing intervention research and development. FPKM values are defined as: Fragments Per Kilobase of coding sequence per Million mapped fragments, and are the units for the expression values given by the differential gene expression analysis program, Cufflinks.