Project Summary
Exploiting Antibiotic Resistance Mechanisms to Combat Antibiotic Resistance
- Principle Investigator(s):
- Douglas R. Call, Lisa H. Orfe, Owen Strom
- Institution(s):
- Washington State University
- Completion Date:
- May 2014
Background
Antimicrobial resistance is a global challenge and food animal production, including cattle production, is increasingly singled‐out as a significant contributor to emergence, amplification, persistence and dissemination of antimicrobial‐resistant bacteria. Regulatory controls and prudent‐use practices are the primary strategies used to control resistance, and these are probably very important to limit the emergence of resistance to new antibiotics. Once the prevalence of resistance has increased to a clinically relevant level, however, it is unclear how effective prudent use and regulatory control practices will be in the short‐term. This is primarily due to the fact that prudent use and regulatory controls rely on a very simple, but ineffective mechanism of “passive decay.” This refers to the process of removing antibiotic selection pressure and “hoping” that the resistant bacteria decline in prevalence. This is likely to happen eventually, but the rate of decline is a function of the fitness cost that is incurred by the bacteria for carrying resistance traits. Unfortunately, many traits cost little to retain in bacterial populations and there are numerous examples in the literature whereby removal of antibiotics has resulted in only limited declines in resistant populations of antibiotic resistant bacteria.
What is needed is strategy to directly select against antibiotic resistant bacteria. Under this scenario, a highly prevalent antibiotic resistance trait would be targeted and a fitness cost on the host bacterium would be induced as the “price” for harboring the resistance trait. The fitness cost would make the bacteria less competitive so that the bacteria would rapidly diminish in prevalence, or they would shed the resistance trait. Unlike passive decline, active selection against antibiotic resistance would happen in a matter of days instead of years. There are several candidate resistance traits to consider for this approach. Tetracycline efflux pumps are of particular interest because of their broad global distribution and because they are regulated so that their expression only occurs in the presence of tetracycline or a tetracycline analogue.
Presumably, this tight regulation evolved to limit the fitness cost for expressing these traits when they are not needed. For the current proposal the researchers of this study developed a means to induce expression of tetracycline efflux pumps. They showed that this method activates the resistance genes while not having any antibacterial activity against susceptible bacteria. Feeding inducer tetracycline resistance to dairy calves produced mixed results, but this has led to several additional ideas for developing this strategy.
The specific objectives of the study were to (1) develop a protocol for preparing a tetracycline‐resistance inducer and confirm activation of efflux pump synthesis without antibacterial function; and (2) conduct a clinical trial to determine if feeding calves the tetracycline inducer will reduce the proportion of tetracycline and multidrug resistant E. coli in cattle feces.
Methodology
The inducer is a commercially available veterinary antibiotic that is modified by “cooking” the antibiotic for a defined temperature and time. Bacterial culture experiments were used to assess the impact of the degraded antibiotic on susceptible and resistant E. coli strains. This involved treating the cultures with modified and unmodified antibiotic and using agar plate counts with selective media to quantify changes in resistant and susceptible populations of E. coli.
Subsequently, RNA was harvested from the bacteria and a molecular assay (real‐time PCR) was used to quantify changes in expression of two tetracycline efflux pump genes. It was originally proposed to test this idea by feeding dairy calves a daily dose of the modified antibiotic and monitor changes in the counts of resistant E. coli in the calf feces. Immediately before the initiation of the trial with 20 calves at a working dairy, the herd veterinarian warned that the experiment would probably be considered an extra‐label use of antibiotics for food animals by the FDA, and the animals might have to be destroyed to prevent their introduction into the food chain.
Consequently, it was necessary to purchase surplus bull calves and conduct the experiment in a controlled environment followed by euthanizing the treated calves. This revised protocol worked, but it severely limited the sample size that could be included for these initial trials.
Findings
Heating the commercial antibiotic produced eight distinct degradation products as determined by HPLC‐UV (not shown). Defined strains of E. coli were exposed to the degraded product (“inducer”) and the sensitive strain of E. coli grew normally (not shown). Exposure to degraded products (two different formulations) induced expression of two tetracycline efflux pump genes (Fig. 1, tet(A); tet(B) not shown). Having satisfied these conditions, three feeding trials were conducted. For trial 1, after 14 days, treated calves had 70‐fold fewer tetracycline-resistant E. coli compared to untreated controls. Furthermore, it was not possible to detect any multi‐drug resistant E. coli, which was expected because most multi‐drug resistant strains harbor tetracycline resistance. Trial 2 was rejected from the study because there were significant calf health problems encountered before conducting the trial. Trial 3 resulted in a non‐significant effect. Pooling the results from trials 1 and 2 showed a 3‐fold combined reduction of tetracycline‐resistant E. coli, but this was not statistically significant. Additional research indicates that for the proposed strategy to work, cofactors needed to be present that take advantage of the fact that these bacteria express resistance genes when they are not needed. Findings from the current study have been leveraged into an intramural research project at Washington State University that will identify cofactors and determine which component of the degraded antibiotic is responsible for inducing expression of the resistance genes.
Implications
Provided that the researchers of this study can use the proposed strategy to actively select against antibiotic resistant bacteria, the implications for the industry are significant. At this time, no other sector in human or veterinary medicine has ever attempted to actively select against resistant bacteria as the researchers of this study have proposed. Consequently, the industry will have the first ever proactive strategy for reducing the prevalence of antibiotic resistant bacteria if successful. This will have both real benefits in terms of animal health and public health while having an important political benefit because it will demonstrate that the industry is genuinely committed to finding solutions to the antibiotic resistance challenge in food animal production.
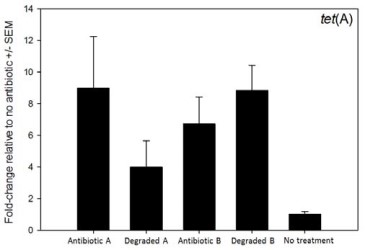
Figure 1. Effect of two degraded antibiotic products on expression of antibiotic resistance gene tet(A). Exposure to active and inactivated antibiotics resulted in increased expression of the resistance gene.